Linear Regression Model in Data Science
To understand how to fit a given set of data into a Linear Regression Model, let’s go through an example.
We will construct a linear model that explains the relationship a car’s mileage (mpg) has with its other attributes
Import Libraries
In [1]:
import numpy as np from sklearn.linear_model import LinearRegression import pandas as pd import matplotlib.pyplot as plt %matplotlib inline import seaborn as sns from sklearn.model_selection import train_test_split # Sklearn package's randomized data splitting function
C:\Users\Rajet singh\Anaconda3\lib\site-packages\statsmodels\tools\_testing.py:19: FutureWarning: pandas.util.testing is deprecated. Use the functions in the public API at pandas.testing instead. import pandas.util.testing as tm
Load and review data
In [2]:
cData = pd.read_csv("auto-mpg.csv")
cData.shape
Out[2]:
(398, 9)
In [3]:
# 8 variables: # # MPG (miles per gallon), # cylinders, # engine displacement (cu. inches), # horsepower, # vehicle weight (lbs.), # time to accelerate from O to 60 mph (sec.), # model year (modulo 100), and # origin of car (1. American, 2. European,3. Japanese). # # Also provided are the car labels (types) # Missing data values are marked by series of question marks. cData.head()
Out[3]:
| mpg | cylinders | displacement | horsepower | weight | acceleration | model year | origin | car name | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 18.0 | 8 | 307.0 | 130 | 3504 | 12.0 | 70 | 1 | chevrolet chevelle malibu |
| 1 | 15.0 | 8 | 350.0 | 165 | 3693 | 11.5 | 70 | 1 | buick skylark 320 |
| 2 | 18.0 | 8 | 318.0 | 150 | 3436 | 11.0 | 70 | 1 | plymouth satellite |
| 3 | 16.0 | 8 | 304.0 | 150 | 3433 | 12.0 | 70 | 1 | amc rebel sst |
| 4 | 17.0 | 8 | 302.0 | 140 | 3449 | 10.5 | 70 | 1 | ford torino |
In [4]:
#dropping/ignoring car_name
cData = cData.drop('car name', axis=1)
# Also replacing the categorical var with actual values
cData['origin'] = cData['origin'].replace({1: 'america', 2: 'europe', 3: 'asia'})
cData.head()
Out[4]:
| mpg | cylinders | displacement | horsepower | weight | acceleration | model year | origin | |
|---|---|---|---|---|---|---|---|---|
| 0 | 18.0 | 8 | 307.0 | 130 | 3504 | 12.0 | 70 | america |
| 1 | 15.0 | 8 | 350.0 | 165 | 3693 | 11.5 | 70 | america |
| 2 | 18.0 | 8 | 318.0 | 150 | 3436 | 11.0 | 70 | america |
| 3 | 16.0 | 8 | 304.0 | 150 | 3433 | 12.0 | 70 | america |
| 4 | 17.0 | 8 | 302.0 | 140 | 3449 | 10.5 | 70 | america |
Create Dummy Variables
Values like ‘america’ cannot be read into an equation. Using substitutes like 1 for america, 2 for europe and 3 for asia would end up implying that european cars fall exactly half way between american and asian cars! we dont want to impose such an baseless assumption!
So we create 3 simple true or false columns with titles equivalent to “Is this car America?”, “Is this care European?” and “Is this car Asian?”. These will be used as independent variables without imposing any kind of ordering between the three regions.
In [5]:
cData = pd.get_dummies(cData, columns=['origin']) cData.head()
Out[5]:
| mpg | cylinders | displacement | horsepower | weight | acceleration | model year | origin_america | origin_asia | origin_europe | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 18.0 | 8 | 307.0 | 130 | 3504 | 12.0 | 70 | 1 | 0 | 0 |
| 1 | 15.0 | 8 | 350.0 | 165 | 3693 | 11.5 | 70 | 1 | 0 | 0 |
| 2 | 18.0 | 8 | 318.0 | 150 | 3436 | 11.0 | 70 | 1 | 0 | 0 |
| 3 | 16.0 | 8 | 304.0 | 150 | 3433 | 12.0 | 70 | 1 | 0 | 0 |
| 4 | 17.0 | 8 | 302.0 | 140 | 3449 | 10.5 | 70 | 1 | 0 | 0 |
Dealing with Missing Values
In [6]:
#A quick summary of the data columns cData.describe()
Out[6]:
| mpg | cylinders | displacement | weight | acceleration | model year | origin_america | origin_asia | origin_europe | |
|---|---|---|---|---|---|---|---|---|---|
| count | 398.000000 | 398.000000 | 398.000000 | 398.000000 | 398.000000 | 398.000000 | 398.000000 | 398.000000 | 398.000000 |
| mean | 23.514573 | 5.454774 | 193.425879 | 2970.424623 | 15.568090 | 76.010050 | 0.625628 | 0.198492 | 0.175879 |
| std | 7.815984 | 1.701004 | 104.269838 | 846.841774 | 2.757689 | 3.697627 | 0.484569 | 0.399367 | 0.381197 |
| min | 9.000000 | 3.000000 | 68.000000 | 1613.000000 | 8.000000 | 70.000000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 17.500000 | 4.000000 | 104.250000 | 2223.750000 | 13.825000 | 73.000000 | 0.000000 | 0.000000 | 0.000000 |
| 50% | 23.000000 | 4.000000 | 148.500000 | 2803.500000 | 15.500000 | 76.000000 | 1.000000 | 0.000000 | 0.000000 |
| 75% | 29.000000 | 8.000000 | 262.000000 | 3608.000000 | 17.175000 | 79.000000 | 1.000000 | 0.000000 | 0.000000 |
| max | 46.600000 | 8.000000 | 455.000000 | 5140.000000 | 24.800000 | 82.000000 | 1.000000 | 1.000000 | 1.000000 |
In [7]:
# hp is missing cause it does not seem to be reqcognized as a numerical column! cData.dtypes
Out[7]:
mpg float64 cylinders int64 displacement float64 horsepower object weight int64 acceleration float64 model year int64 origin_america uint8 origin_asia uint8 origin_europe uint8 dtype: object
In [8]:
# isdigit()? on 'horsepower' hpIsDigit = pd.DataFrame(cData.horsepower.str.isdigit()) # if the string is made of digits store True else False #print isDigit = False! cData[hpIsDigit['horsepower'] == False] # from temp take only those rows where hp has false
Out[8]:
| mpg | cylinders | displacement | horsepower | weight | acceleration | model year | origin_america | origin_asia | origin_europe | |
|---|---|---|---|---|---|---|---|---|---|---|
| 32 | 25.0 | 4 | 98.0 | ? | 2046 | 19.0 | 71 | 1 | 0 | 0 |
| 126 | 21.0 | 6 | 200.0 | ? | 2875 | 17.0 | 74 | 1 | 0 | 0 |
| 330 | 40.9 | 4 | 85.0 | ? | 1835 | 17.3 | 80 | 0 | 0 | 1 |
| 336 | 23.6 | 4 | 140.0 | ? | 2905 | 14.3 | 80 | 1 | 0 | 0 |
| 354 | 34.5 | 4 | 100.0 | ? | 2320 | 15.8 | 81 | 0 | 0 | 1 |
| 374 | 23.0 | 4 | 151.0 | ? | 3035 | 20.5 | 82 | 1 | 0 | 0 |
In [9]:
# Missing values have a'?''
# Replace missing values with NaN
cData = cData.replace('?', np.nan)
cData[hpIsDigit['horsepower'] == False]
Out[9]:
| mpg | cylinders | displacement | horsepower | weight | acceleration | model year | origin_america | origin_asia | origin_europe | |
|---|---|---|---|---|---|---|---|---|---|---|
| 32 | 25.0 | 4 | 98.0 | NaN | 2046 | 19.0 | 71 | 1 | 0 | 0 |
| 126 | 21.0 | 6 | 200.0 | NaN | 2875 | 17.0 | 74 | 1 | 0 | 0 |
| 330 | 40.9 | 4 | 85.0 | NaN | 1835 | 17.3 | 80 | 0 | 0 | 1 |
| 336 | 23.6 | 4 | 140.0 | NaN | 2905 | 14.3 | 80 | 1 | 0 | 0 |
| 354 | 34.5 | 4 | 100.0 | NaN | 2320 | 15.8 | 81 | 0 | 0 | 1 |
| 374 | 23.0 | 4 | 151.0 | NaN | 3035 | 20.5 | 82 | 1 | 0 | 0 |
There are various ways to handle missing values. Drop the rows, replace missing values with median values etc. of the 398 rows 6 have NAN in the hp column. We could drop those 6 rows – which might not be a good idea under all situations
In [10]:
#instead of dropping the rows, lets replace the missing values with median value. cData.median()
Out[10]:
mpg 23.0 cylinders 4.0 displacement 148.5 horsepower 93.5 weight 2803.5 acceleration 15.5 model year 76.0 origin_america 1.0 origin_asia 0.0 origin_europe 0.0 dtype: float64
In [11]:
# replace the missing values with median value.
# Note, we do not need to specify the column names below
# every column's missing value is replaced with that column's median respectively (axis =0 means columnwise)
#cData = cData.fillna(cData.median())
medianFiller = lambda x: x.fillna(x.median())
cData = cData.apply(medianFiller,axis=0)
cData['horsepower'] = cData['horsepower'].astype('float64') # converting the hp column from object / string type to float
BiVariate Plots
A bivariate analysis among the different variables can be done using scatter matrix plot. Seaborn libs create a dashboard reflecting useful information about the dimensions. The result can be stored as a .png file.
In [12]:
cData_attr = cData.iloc[:, 0:7] sns.pairplot(cData_attr, diag_kind='kde') # to plot density curve instead of histogram on the diag
Out[12]:
<seaborn.axisgrid.PairGrid at 0x2aba2117108>
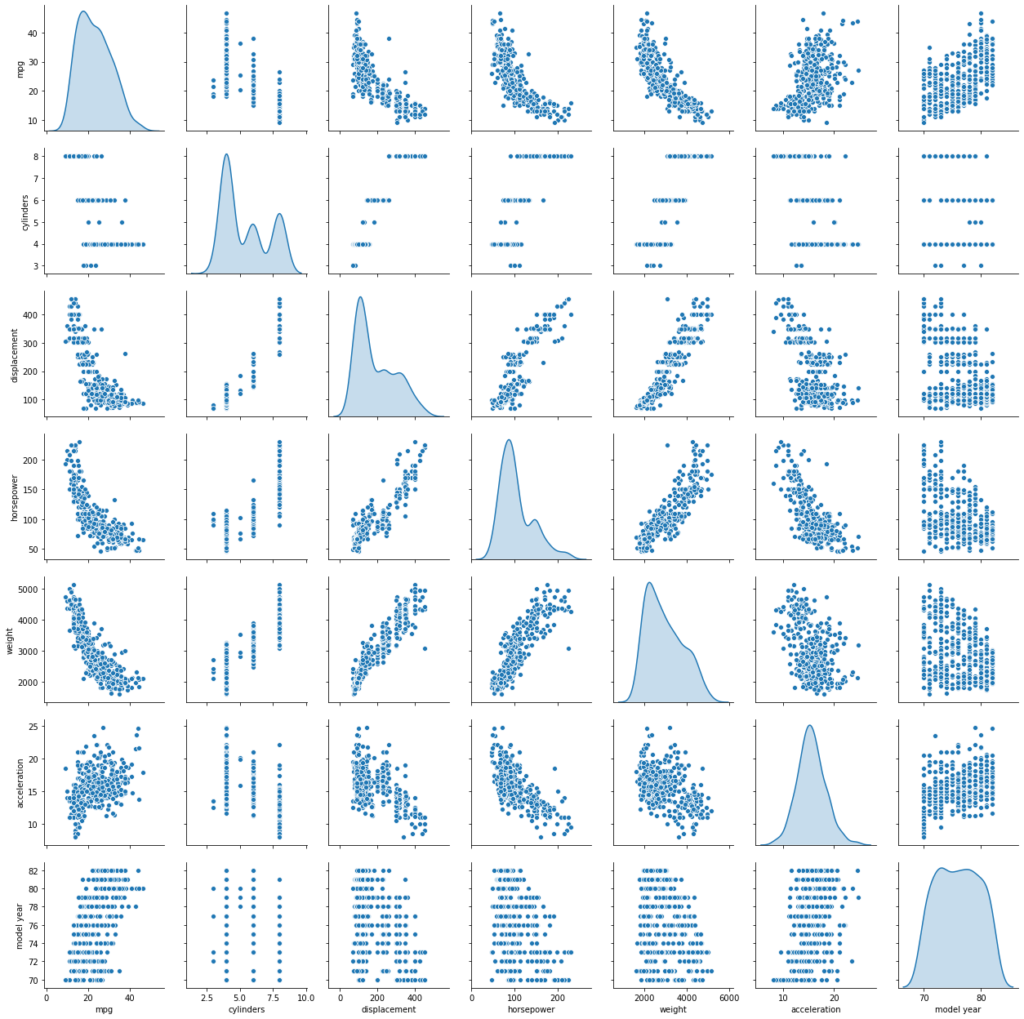
Observation between ‘mpg’ and other attributes indicate the relationship is not really linear. However, the plots also indicate that linearity would still capture quite a bit of useful information/pattern. Several assumptions of classical linear regression seem to be violated, including the assumption of no Heteroscedasticity
Split Data
In [13]:
# lets build our linear model # independant variables X = cData.drop(['mpg','origin_europe'], axis=1) # the dependent variable y = cData[['mpg']]
In [14]:
# Split X and y into training and test set in 70:30 ratio X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=1)
Fit Linear Model
In [15]:
regression_model = LinearRegression() regression_model.fit(X_train, y_train)
Out[15]:
LinearRegression(copy_X=True, fit_intercept=True, n_jobs=None, normalize=False)
Here are the coefficients for each variable and the intercept
In [16]:
for idx, col_name in enumerate(X_train.columns):
print("The coefficient for {} is {}".format(col_name, regression_model.coef_[0][idx]))
The coefficient for cylinders is -0.3948079661648244 The coefficient for displacement is 0.028945510765487174 The coefficient for horsepower is -0.02175220772354676 The coefficient for weight is -0.007352032065147351 The coefficient for acceleration is 0.061919366007618326 The coefficient for model year is 0.8369338917644993 The coefficient for origin_america is -3.0012830009185123 The coefficient for origin_asia is -0.6060179643247369
In [17]:
intercept = regression_model.intercept_[0]
print("The intercept for our model is {}".format(intercept))
The intercept for our model is -18.283451116372067
The score (R^2) for in-sample and out of sample
In [18]:
regression_model.score(X_train, y_train)
Out[18]:
0.8141025501610559
In [19]:
#out of sample score (R^2) regression_model.score(X_test, y_test)
Out[19]:
0.8433135132808833
Adding interaction terms
In [20]:
from sklearn.preprocessing import PolynomialFeatures from sklearn import linear_model poly = PolynomialFeatures(degree=2, interaction_only=True) X_train2 = poly.fit_transform(X_train) X_test2 = poly.fit_transform(X_test) poly_clf = linear_model.LinearRegression() poly_clf.fit(X_train2, y_train) y_pred = poly_clf.predict(X_test2) #print(y_pred) #In sample (training) R^2 will always improve with the number of variables! print(poly_clf.score(X_train2, y_train))
0.9015975294607972
In [21]:
#Out off sample (testing) R^2 is our measure of sucess and does improve print(poly_clf.score(X_test2, y_test))
0.8647441061208313
In [22]:
# but this improves as the cost of 29 extra variables! print(X_train.shape) print(X_train2.shape)
(278, 8) (278, 37)
Polynomial Features (with only interaction terms) have improved the Out of sample R^2. However at the cost of increaing the number of variables significantly.